UChicago CS Researchers Share in Special Prize on COVID-19 Research
When the COVID-19 pandemic began, scientists of all disciplines rushed to apply their skills and knowledge to the global crisis. That included computer scientists at the University of Chicago and Argonne National Laboratory, who joined an international collaboration to adapt AI language models for discovering variants of the SARS-CoV-2 virus and the potential for new more dangerous or transmissible forms.
At this year’s International Conference for High Performance Computing, Networking, Storage, and Analysis (SC22), that collaboration was given the ACM Gordon Bell Special Prize for HPC-Based COVID-19 Research. The paper, “GenSLMs: Genome-Scale Language Models Reveal SARS-CoV-2 Evolutionary Dynamics,” was honored for outstanding research achievement toward the understanding of the COVID-19 pandemic through the use of high-performance computing.

UChicago authors on the paper included Professors of Computer Science Ian Foster and Rick Stevens, PhD students Alexander Brace, Austin Clyde, and J. Gregory Pauloski, undergraduate student Diangen (Dana) Lin, and postdoctoral researcher Valerie Hayot-Sasson. The collaboration also included several scientists from our partner Argonne National Laboratory, who wrote a feature piece on the award-winning research:
“When the pandemic began, we had several of these really harmful variants of the virus, like the Delta variant,” said Argonne computational biologist Arvind Ramanathan. “It resulted in a large death toll. But Delta evolved as a consequence of certain mutations that were happening when the virus was facing the human hosts. It’s a process of evolution of the virus inside of the human cell.”
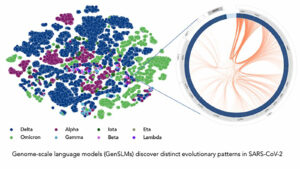
Their work resulted in the first genome-scale language model (GenSLM), which is a model that can analyze genes and rapidly identify VOCs. The model discussed in the paper was trained on data from the COVID-19 pandemic, and the hope is that models like this could potentially give health officials the tools they need to quickly respond to rising variants. GenSLM is the first whole genome-scale foundation model that can be altered and applied to other prediction tasks similar to VOC identification.
While these evolutionary variants may seem to crop up randomly to the human eye, tracking them is of the utmost concern. As such, the work of Ramanathan and his colleagues could seriously alter how we stay on top of viral outbreaks.
Read more about the project and how it utilized Argonne Leadership Computing Facility resources such as the Polaris supercomputer and the ALCF AI Testbed.












