UChicago Team Wins The NIH Long COVID Computational Challenge
COVID-19 has plagued the world since early 2020, infecting over three quarters of a billion people. However, there is still plenty that scientists do not know about how the virus will manifest in patients. One of the most pressing unanswered questions right now is how to predict which patients will experience Long COVID: broadly defined by the CDC as signs, symptoms, or conditions that persist or develop after an initial infection. When a competition arose calling for data-driven solutions that meaningfully advance the current understanding of risks for developing Long COVID, a team of scientists from the University of Chicago’s Department of Computer Science and Argonne National Laboratory offered a solution worth the $200,000 grand prize.
The Rapid Acceleration of Diagnostics Radical program at the National Institutes of Health (NIH) launched the Long COVID Computational Challenge in August of 2022. The primary goal was to generate the development of AI and machine learning algorithms that can be used as an open-source tool to identify which patients that had previously contracted COVID-19 would be most likely to develop Long Covid. Participants received a set of de-identified electronic health record (EHR) data that was representative of 74 health centers across the United States. After developing, training, and testing their models, submissions went through a quantitative and qualitative evaluation by a panel of federal researchers. A total of 74 teams, consisting of over 300 scientists from universities, medical centers, and industry leaders, registered to compete. Out of those 74, only 35 teams completed submissions.

First place team, Convalesco, included UChicago third-year Ph.D. student Zixuan Zhao, CS alumnus and Computer Scientist Fangfang Xia (Argonne), and Computational Scientist Yitan Zhu (Argonne).
“None of us is a domain expert in this problem,” said Xia. “I would say we have some experience applying machine learning to medical problems, but we could have benefited from having doctors and statisticians on the team. However, I think that probably also gives us a fresh eye to look at the data for what it is. We spent a lot of time just looking at the data and doing qualitative exploration.”
The group built a lightweight monitoring system using machine learning models that updates a patient’s likelihood for developing Long COVID in real time as new symptoms are reported. Part of their technological challenge was dealing with the irregularity of medical data.
“A lot of machine learning progress has been made in images and language, but medical data can be kind of messy,” explained Xia. “For one, we have diverse data modalities including medical procedures, drug exposures, lab measurements, and symptoms that are often called by many different aliases. Deep learning models don’t deal well with tabular data of this nature. Moreover, all these features are time course data with irregular timestamps. For instance, a patient may not go to the doctor for years and then suddenly has frequent visits. That kind of data is always a challenge, but it’s also typical of many real-world applications. So part of our focus was developing feature engineering methods for these types of timed events.”
Another focus was making sure the model was generalizable.
“We were always conscious of how the model might be tested or put to use in real life, so we tried to remove features instead of adding more. Narrowing down the data points from thousands of possible symptoms within the patient records allowed us to key in on the critical feature categories that are most likely playing a role in the disease.”
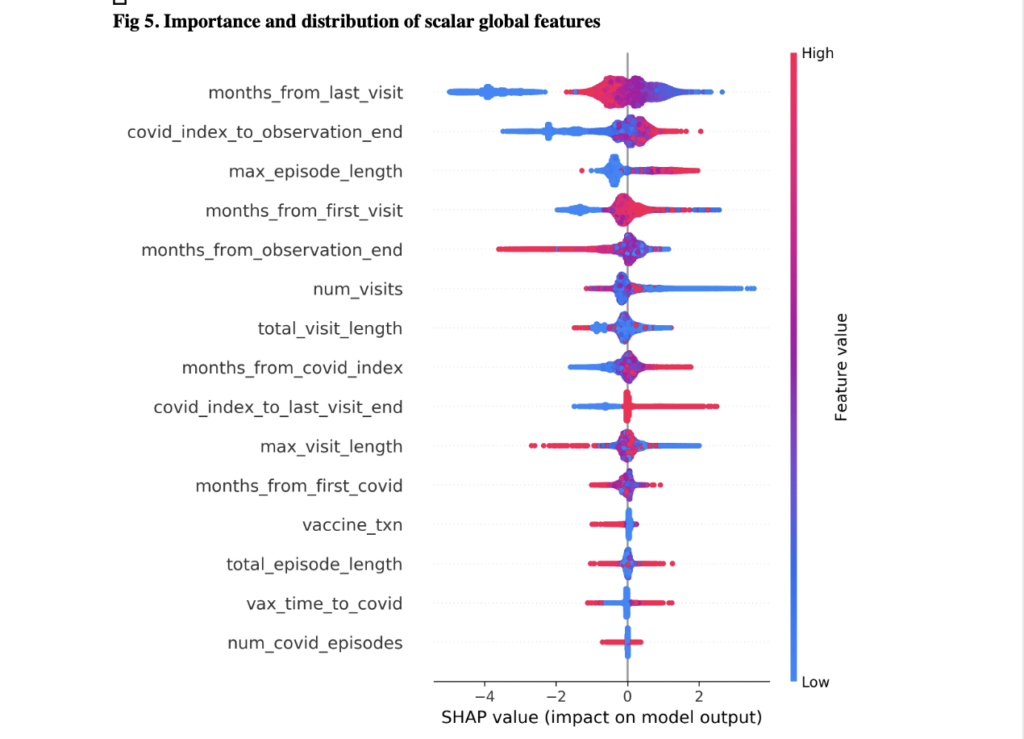 Overall, their visualization dashboard utilized 100 symptoms and 31 demographic features within the data to build a timeline of the cumulative risk. Some of those features were conditions from the acute phase (such as fatigue, pain, and weakness) as well as oxygen saturation, certain drug exposures, and bloodwork. For a real world application, doctors could first look at a patient’s demographic information and gather some sense of a base risk of this person developing Long COVID. Then they would look at medical records before COVID and input other diseases that would increase a patient’s risk. Finally, doctors would enter information gathered over a 28 day period while the patient is being monitored. As all of these measurements are being entered, the visualization dashboard is showing the likelihood of that patient developing Long COVID three months, six months, or even a year down the road. This would help doctors determine whether to undergo different treatment early on.
Overall, their visualization dashboard utilized 100 symptoms and 31 demographic features within the data to build a timeline of the cumulative risk. Some of those features were conditions from the acute phase (such as fatigue, pain, and weakness) as well as oxygen saturation, certain drug exposures, and bloodwork. For a real world application, doctors could first look at a patient’s demographic information and gather some sense of a base risk of this person developing Long COVID. Then they would look at medical records before COVID and input other diseases that would increase a patient’s risk. Finally, doctors would enter information gathered over a 28 day period while the patient is being monitored. As all of these measurements are being entered, the visualization dashboard is showing the likelihood of that patient developing Long COVID three months, six months, or even a year down the road. This would help doctors determine whether to undergo different treatment early on.
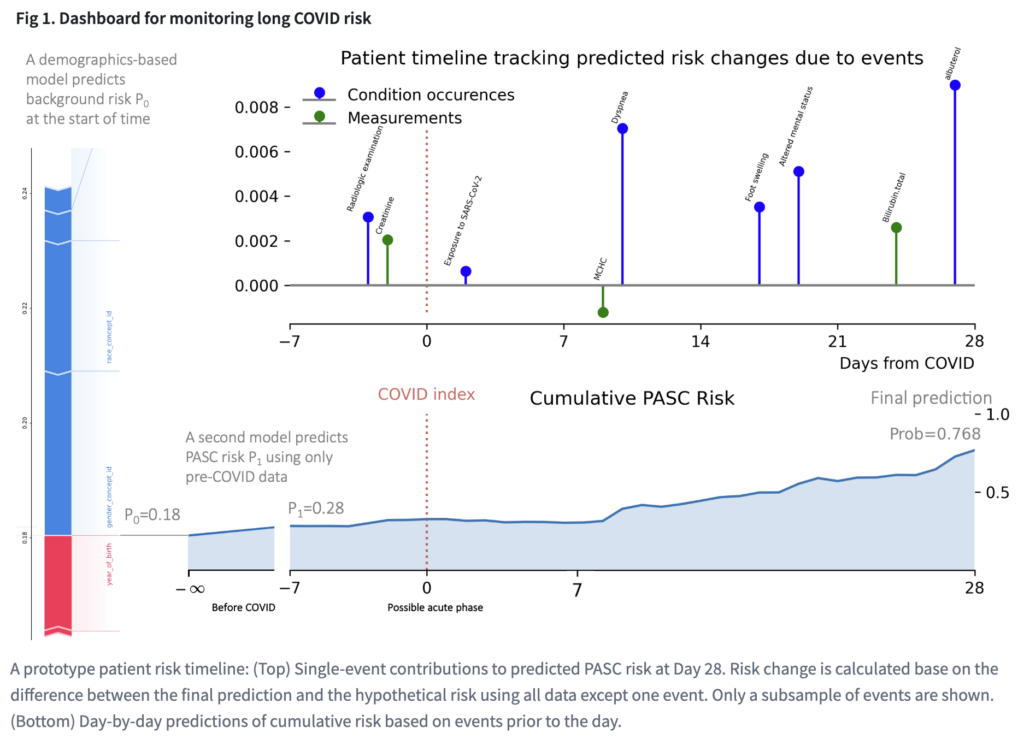
During the competition, the model was tested by applying those features to the medical records of a large group of patient’s EHR data, both before COVID was diagnosed and once there was an onset. The team was not able to use records that surpassed 28 days for training, but they were told whether that patient eventually developed Long COVID. The ultimate task was then using this information and their model to predict whether other patients would develop Long COVID down the line.
Once teams submitted their models, it was tested against similar data withheld during the entry phase. Months later, it was tested against data from new patients in new locations to see if the model held up. Convalesco was the only team to improve their accuracy from the first set of data to the second.
“We are exploring ways to make these models even more generalizable,” said Zhao. “One of my research interests is self-supervised learning, which is like learning without a teacher. This could reduce the need for costly manual labels, and perhaps equally useful, it forces the model to use the information in the data itself to learn a better, more coherent representation. We have seen its success in vision and language models. For example, these models can learn by recovering a rotated image or predicting the next word. However, such self-supervision targets are much less obvious in medical data and many scientific domains. This motivates us to look for novel and perhaps more general learning algorithms.”
Although the team doesn’t know whether their model will be implemented in the medical field, they are happy to allow others the chance to extend upon their work. The full code was released in the open source world and can be accessed by anyone. They hope it can represent a class of problems within medical data that still need to be solved. Xia believes that making progress in AI boils down to conviction.
“An interesting psychological effect in many fields is that once you know something is doable, it suddenly becomes achievable for everyone. Short of knowing, believing may be the next best. This is why we appreciate competitions like this: it invites you to think hard about what’s possible. Now, the next conviction we are looking to test is how much of what we, data scientists, do can be automated. In theory, our lack of domain expertise should make it easier for models to emulate us, but we have tried and have not yet succeeded in replacing ourselves. That means there is still some elusive insight in our workflow that we need to capture.”














